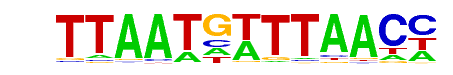
Hnf1(Homeobox)/Liver-Foxa2-Chip-Seq(GSE25694)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTTAAACATTAA
GGTTAAACATTAA |
|


|
|
MA0153.1_HNF1B/Jaspar
| Match Rank: | 2 |
| Score: | 0.93
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA
-GTTAAATATTAA |
|


|
|
MA0046.1_HNF1A/Jaspar
| Match Rank: | 3 |
| Score: | 0.84
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGTTAAACATTAA-
GGTTAATAATTAAC |
|


|
|
HAHB4(HD-ZIP)/Helianthus anuus/AthaMap
| Match Rank: | 4 |
| Score: | 0.71
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA
---CAATCATTA- |
|


|
|
Dfd/dmmpmm(Bigfoot)/fly
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA
-----ATAATTAA |
|


|
|
Dfd/dmmpmm(Pollard)/fly
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA
-----ATAATTAA |
|


|
|
br-Z4/dmmpmm(SeSiMCMC)/fly
| Match Rank: | 7 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | GGTTAAACATTAA
---TAATTATT-- |
|


|
|
MA0166.1_Antp/Jaspar
| Match Rank: | 8 |
| Score: | 0.66
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA
------TCATTAA |
|


|
|
PHO2(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.66
| | Offset: | 8
| | Orientation: | forward strand |
| Alignment: | GGTTAAACATTAA-
--------ATTAAG |
|


|
|
eve/dmmpmm(Pollard)/fly
| Match Rank: | 10 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | GGTTAAACATTAA--
---AAANNATTAACG |
|


|
|