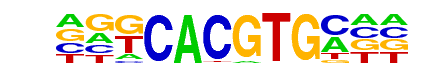HIF2a(HLH)/785_O-HIF2a-ChIP-Seq(GSE34871)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCACGTACCC
GCACGTACCC |
|


|
|
HIF-1a(HLH)/MCF7-HIF1a-ChIP-Seq(GSE28352)/Homer
| Match Rank: | 2 |
| Score: | 0.96
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GCACGTACCC
BGCACGTA--- |
|


|
|
MA0259.1_HIF1A::ARNT/Jaspar
| Match Rank: | 3 |
| Score: | 0.95
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCACGTACCC
GCACGTNC-- |
|


|
|
MA0004.1_Arnt/Jaspar
| Match Rank: | 4 |
| Score: | 0.83
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCACGTACCC
-CACGTG--- |
|


|
|
MA0310.1_HAC1/Jaspar
| Match Rank: | 5 |
| Score: | 0.83
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GCACGTACCC
GACACGTG--- |
|


|
|
MA0281.1_CBF1/Jaspar
| Match Rank: | 6 |
| Score: | 0.82
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCACGTACCC
TCACGTGC-- |
|


|
|
MA0569.1_MYC4/Jaspar
| Match Rank: | 7 |
| Score: | 0.82
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCACGTACCC
GCACGTGN-- |
|


|
|
bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 8 |
| Score: | 0.81
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GCACGTACCC
NGKCACGTGM-- |
|

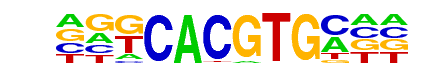
|
|
MA0566.1_MYC2/Jaspar
| Match Rank: | 9 |
| Score: | 0.80
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCACGTACCC
GCACGTGC-- |
|


|
|
OsbHLH66(bHLH)/Oryza sativa/AthaMap
| Match Rank: | 10 |
| Score: | 0.80
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GCACGTACCC-
GAGCACGTGCGCA |
|


|
|