FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCANTGACCTN
AGGTCANTGACCTN |
|


|
|
EcR/usp/dmmpmm(Bergman)/fly
| Match Rank: | 2 |
| Score: | 0.93
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGGTCANTGACCTN
GAGGTCAATGACCTC |
|


|
|
MA0534.1_EcR::usp/Jaspar
| Match Rank: | 3 |
| Score: | 0.83
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGGTCANTGACCTN
AAGGTCAATGAACTN |
|


|
|
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCANTGACCTN
AGGTCA-------- |
|


|
|
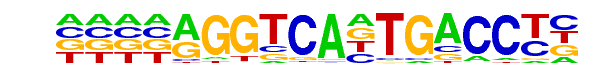
MA0071.1_RORA_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.64
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGGTCANTGACCTN
ATCAAGGTCA-------- |
|

|
|
MA0160.1_NR4A2/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | AGGTCANTGACCTN
------GTGACCTT |
|


|
|
Hr46/dmmpmm(Bergman)/fly
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCANTGACCTN
GGGTCA-------- |
|


|
|
Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 8 |
| Score: | 0.62
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | AGGTCANTGACCTN--
------NTGACCTTGA |
|


|
|
RARg(NR)/ES-RARg-ChIP-Seq(GSE30538)/Homer
| Match Rank: | 9 |
| Score: | 0.61
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCANTGACCTN
AGGTCAAGGTCA-- |
|


|
|
RTG3/Literature(Harbison)/Yeast
| Match Rank: | 10 |
| Score: | 0.61
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | AGGTCANTGACCTN
------GTGACC-- |
|


|
|





















