ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT
ANCCGGAAGT |
|


|
|
ETS(ETS)/Promoter/Homer
| Match Rank: | 2 |
| Score: | 0.99
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT
AACCGGAAGT |
|


|
|
GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 3 |
| Score: | 0.98
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT
NACCGGAAGT |
|


|
|
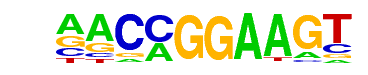
ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer
| Match Rank: | 4 |
| Score: | 0.97
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT
AACCGGAAGT |
|

|
|
Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.96
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ANCCGGAAGT-
-RCCGGAAGTD |
|


|
|
MA0076.2_ELK4/Jaspar
| Match Rank: | 6 |
| Score: | 0.96
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ANCCGGAAGT--
-NCCGGAAGTGG |
|


|
|
MA0062.2_GABPA/Jaspar
| Match Rank: | 7 |
| Score: | 0.96
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT---
--CCGGAAGTGGC |
|


|
|
PB0011.1_Ehf_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.96
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ANCCGGAAGT--
AGGACCCGGAAGTAA |
|


|
|
Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 9 |
| Score: | 0.94
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ANCCGGAAGT-
-RCCGGAARYN |
|


|
|
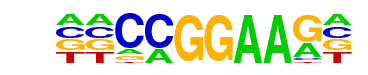
MA0026.1_Eip74EF/Jaspar
| Match Rank: | 10 |
| Score: | 0.94
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ANCCGGAAGT
--CCGGAAG- |
|

|
|





















