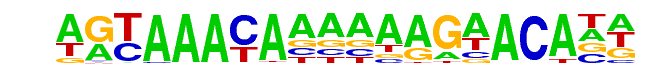
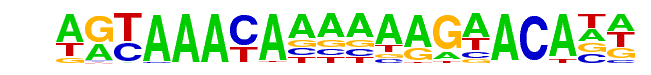
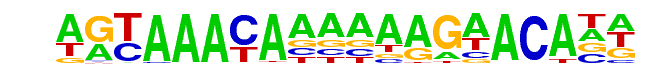
FOXA1:AR/LNCAP-AR-ChIP-Seq(GSE27824)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGTAAACAAAAAAGAACANN
AGTAAACAAAAAAGAACANN |
|
|
|
PB0016.1_Foxj1_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGTAAACAAAAAAGAACANN
AAAGTAAACAAAAATT------ |
|

|
|
MA0593.1_FOXP2/Jaspar
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGTAAACAAAAAAGAACANN
AAGTAAACAAA---------- |
|


|
|
MF0005.1_Forkhead_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGTAAACAAAAAAGAACANN
AAATAAACA------------ |
|


|
|
PHA-4(Forkhead)/cElegans-Embryos-PHA4-ChIP-Seq(modEncode)/Homer
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGTAAACAAAAAAGAACANN
-GCAAACAM----------- |
|

|
|
MA0296.1_FKH1/Jaspar
| Match Rank: | 6 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----AGTAAACAAAAAAGAACANN
TGAATTGTAAACAAAGAGGA----- |
|


|
|
MA0481.1_FOXP1/Jaspar
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGTAAACAAAAAAGAACANN
CAAAAGTAAACAAAG--------- |
|


|
|
PB0015.1_Foxa2_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGTAAACAAAAAAGAACANN
AAAAAGTAAACAAAGAC------- |
|


|
|
FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGTAAACAAAAAAGAACANN
NDGTAAACARRN--------- |
|


|
|
FKH1/FKH1_YPD/53-FKH1,53-FKH2(Harbison)/Yeast
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGTAAACAAAAAAGAACANN
-GTAAACAA----------- |
|

|
|