E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | VDTTTCCCGCCA
VDTTTCCCGCCA |
|


|
|
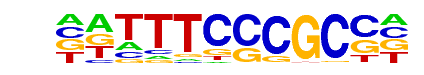
E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 2 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA
-DTTTCCCGCC- |
|

|
|
E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer
| Match Rank: | 3 |
| Score: | 0.96
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA-
---TTCCCGCCWG |
|


|
|
MA0470.1_E2F4/Jaspar
| Match Rank: | 4 |
| Score: | 0.96
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA
-NNTTCCCGCCC |
|


|
|
E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.95
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA
-NYTTCCCGCC- |
|


|
|
MA0471.1_E2F6/Jaspar
| Match Rank: | 6 |
| Score: | 0.92
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA
-NCTTCCCGCCC |
|


|
|
MA0541.1_EFL-1/Jaspar
| Match Rank: | 7 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA---
AATTTCGCGCGNNNN |
|


|
|
MA0024.2_E2F1/Jaspar
| Match Rank: | 8 |
| Score: | 0.90
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA-
--CCTCCCGCCCN |
|


|
|
E2F(E2F)/Cell-Cycle-Exp/Homer
| Match Rank: | 9 |
| Score: | 0.88
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | VDTTTCCCGCCA-
-TTTTCGCGCGAA |
|


|
|
MA0469.1_E2F3/Jaspar
| Match Rank: | 10 |
| Score: | 0.87
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | VDTTTCCCGCCA------
---CTCCCGCCCCCACTC |
|


|
|





















