AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATGCCCTGAGGC
ATGCCCTGAGGC |
|


|
|
AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 2 |
| Score: | 0.99
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC
WTGSCCTSAGGS |
|


|
|
MA0003.2_TFAP2A/Jaspar
| Match Rank: | 3 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC----
-TGCCCTGAGGCANTN |
|


|
|
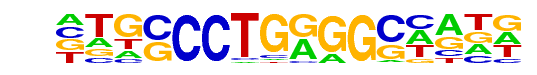
MA0524.1_TFAP2C/Jaspar
| Match Rank: | 4 |
| Score: | 0.93
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC----
-TGCCCTGGGGCNANN |
|

|
|
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.90
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC---
NTNGCCTCAGGCNNN |
|


|
|
PB0087.1_Tcfap2c_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.90
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC---
NTCGCCTCAGGCAAT |
|


|
|
PB0085.1_Tcfap2a_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.88
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC---
NTCCCCTCAGGGANT |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.88
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC--
NTGCCCTAGGGCAA |
|


|
|
PB0189.1_Tcfap2a_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.82
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC--
NTGCCCANNGGTNA |
|


|
|
PB0191.1_Tcfap2c_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.77
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGCCCTGAGGC--
NTGCCCTTGGGCGN |
|


|
|





















