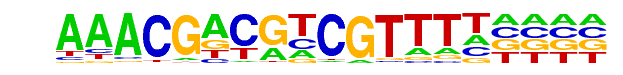
Unknown2/Arabidopsis-Promoters/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AAACGACGTCGTTTT
AAACGACGTCGTTTT |
|


|
|
PB0131.1_Gmeb1_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AAACGACGTCGTTTT
TGGGCGACGTCGTTAA |
|


|
|
MA0411.1_UPC2/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 8
| | Orientation: | reverse strand |
| Alignment: | AAACGACGTCGTTTT
--------TCGTATA |
|


|
|
PH0044.1_Homez/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | AAACGACGTCGTTTT----
--AAAACATCGTTTTTAAG |
|

|
|
MA0395.1_STP2/Jaspar
| Match Rank: | 5 |
| Score: | 0.53
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAACGACGTCGTTTT
NNGNNGTGCGGCGCCGNTNN |
|


|
|
bZIP911(2)(bZIP)/Antirrhinum majus/AthaMap
| Match Rank: | 6 |
| Score: | 0.53
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AAACGACGTCGTTTT
GTACACGTCATC----- |
|


|
|
PB0179.1_Sp100_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.52
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----AAACGACGTCGTTTT
NNTTTANNCGACGNA----- |
|


|
|
STF1(bZIP)/Glycine max/AthaMap
| Match Rank: | 8 |
| Score: | 0.52
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AAACGACGTCGTTTT
-AATGACGTCATT-- |
|


|
|
Adf1/dmmpmm(Pollard)/fly
| Match Rank: | 9 |
| Score: | 0.52
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | AAACGACGTCGTTTT
---CGACCGCG---- |
|


|
|
Unknown4/Arabidopsis-Promoters/Homer
| Match Rank: | 10 |
| Score: | 0.51
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AAACGACGTCGTTTT
---CKTCKTCTTY-- |
|


|
|





















