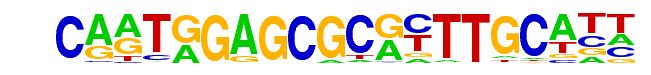
LIN-15B(Zf)/cElegans-L3-LIN15B-ChIP-Seq(modEncode)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
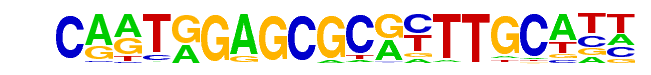
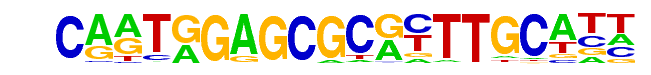
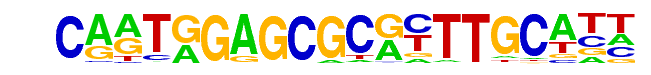
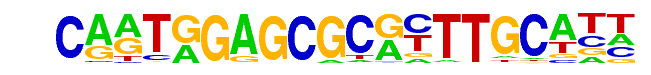
| Alignment: | CARTGGAGCGCRYTTGCATT
CARTGGAGCGCRYTTGCATT |
|
|
|
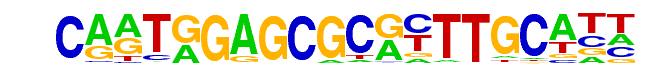
EFL-1(E2F)/cElegans-L1-EFL1-ChIP-Seq(modEncode)/Homer
| Match Rank: | 2 |
| Score: | 0.93
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
---TRGAGCGCRYTTGCA-- |
|

|
|
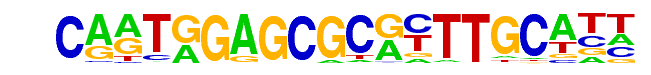
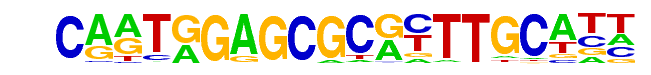
PB0008.1_E2F2_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
--NTCGCGCGCCTTNNN--- |
|

|
|
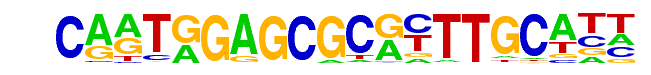
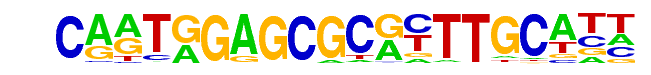
kni/dmmpmm(Pollard)/fly
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
-----GATCTCATTT----- |
|

|
|
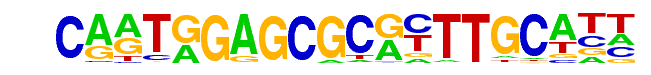
DPL-1(E2F)/cElegans-Adult-ChIP-Seq(modEncode)/Homer
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
---TAGCGCGC--------- |
|

|
|
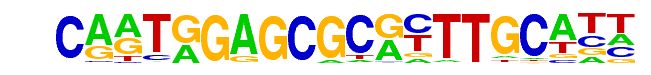
MA0538.1_DAF-12/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
-----GTGTGTGTGTGCGTG |
|

|
|
PB0009.1_E2F3_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
--ANCGCGCGCCCTTNN--- |
|

|
|
MA0291.1_DAL82/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 7
| | Orientation: | reverse strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
-------GCGCACATT---- |
|

|
|
MA0405.1_TEA1/Jaspar
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
-------GCGGACAT----- |
|

|
|
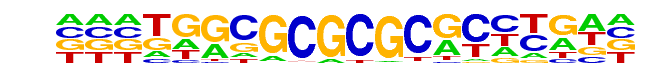
PB0095.1_Zfp161_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CARTGGAGCGCRYTTGCATT
---TGGCGCGCGCGCCTGA- |
|
|
|