Unknown1(NR/Ini-like)/Drosophila-Promoters/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG
MYGGTCACACTG |
|


|
|
RTG3/Literature(Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.79
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG
--GGTCAC---- |
|


|
|
MET31(MacIsaac)/Yeast
| Match Rank: | 3 |
| Score: | 0.71
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | MYGGTCACACTG
---GCCACACC- |
|


|
|
MA0258.2_ESR2/Jaspar
| Match Rank: | 4 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG----
-AGGTCACCCTGACCT |
|


|
|
PCF2(TCP)/Oryza sativa/AthaMap
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG
TGGGGCCCAC-- |
|


|
|
MA0160.1_NR4A2/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG
AAGGTCAC---- |
|


|
|
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | MYGGTCACACTG
-AGGTCA----- |
|


|
|
ERE(NR/IR3)/MCF7-ERa-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | MYGGTCACACTG-----
--GGTCANNGTGACCTN |
|


|
|
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 9 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | MYGGTCACACTG
--KTTCACACCT |
|


|
|
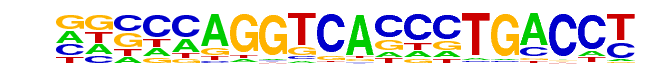
MA0112.2_ESR1/Jaspar
| Match Rank: | 10 |
| Score: | 0.63
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----MYGGTCACACTG----
GGCCCAGGTCACCCTGACCT |
|

|
|





















