E-box/Drosophila-Promoters/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AACAGCTGTTHN
AACAGCTGTTHN |
|


|
|
Ap4(HLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer
| Match Rank: | 2 |
| Score: | 0.96
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AACAGCTGTTHN
-HCAGCTGDTN- |
|


|
|
Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer
| Match Rank: | 3 |
| Score: | 0.95
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AACAGCTGTTHN
-ACAGCTGTTV- |
|


|
|
MyoG(HLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer
| Match Rank: | 4 |
| Score: | 0.95
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | AACAGCTGTTHN
--CAGCTGTT-- |
|


|
|
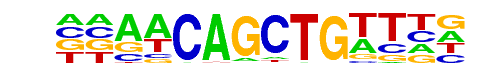
HLH-1(bHLH)/cElegans-Embryo-HLH1-ChIP-Seq(modEncode)/Homer
| Match Rank: | 5 |
| Score: | 0.93
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AACAGCTGTTHN
DVAACAGCTGTY-- |
|

|
|
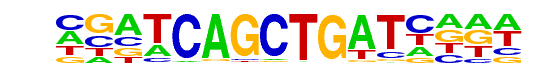
PL0001.1_hlh-11/Jaspar
| Match Rank: | 6 |
| Score: | 0.93
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AACAGCTGTTHN--
NGATCAGCTGATCNNN |
|

|
|
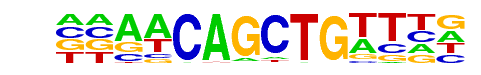
MyoD(HLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer
| Match Rank: | 7 |
| Score: | 0.93
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --AACAGCTGTTHN
NNAGCAGCTGCT-- |
|

|
|
Ptf1a(HLH)/Panc1-Ptf1a-ChIP-Seq(GSE47459)/Homer
| Match Rank: | 8 |
| Score: | 0.92
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AACAGCTGTTHN
-ACAGCTGTTN- |
|


|
|
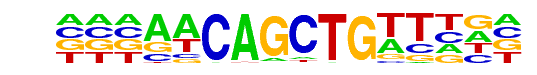
PL0006.1_hlh-2::lin-32/Jaspar
| Match Rank: | 9 |
| Score: | 0.92
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AACAGCTGTTHN-
NNNAACAGCTGTTNNN |
|

|
|
MA0545.1_HLH-1/Jaspar
| Match Rank: | 10 |
| Score: | 0.92
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AACAGCTGTTHN
GNCAGCTGTTN- |
|


|
|





















