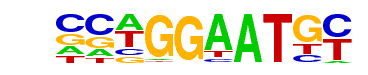
TEAD4(TEA)/Tropoblast-Tead4-ChIP-Seq(GSE37350)/Homer
| Match Rank: | 1 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCWGGAATGY
CCWGGAATGY |
|

|
|
TEAD(TEA)/Fibroblast-PU.1-ChIP-Seq(Unpublished)/Homer
| Match Rank: | 2 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCWGGAATGY
NCTGGAATGC |
|


|
|
MA0090.1_TEAD1/Jaspar
| Match Rank: | 3 |
| Score: | 0.89
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCWGGAATGY-
CNGAGGAATGTG |
|


|
|
MA0406.1_TEC1/Jaspar
| Match Rank: | 4 |
| Score: | 0.87
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CCWGGAATGY
--GGGAATGT |
|


|
|
TEC1/TEC1_YPD/[](Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.85
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCWGGAATGY
--AGGAATG- |
|


|
|
TEC1(MacIsaac)/Yeast
| Match Rank: | 6 |
| Score: | 0.82
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CCWGGAATGY-
---AGAATGTG |
|


|
|
sd/dmmpmm(Pollard)/fly
| Match Rank: | 7 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCWGGAATGY-
CCGAGGAATGTC |
|


|
|
MA0243.1_sd/Jaspar
| Match Rank: | 8 |
| Score: | 0.75
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CCWGGAATGY-
NCGANGAATGTC |
|


|
|
MA0325.1_LYS14/Jaspar
| Match Rank: | 9 |
| Score: | 0.75
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CCWGGAATGY
-CCGGAATT- |
|


|
|
Ik-1
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CCWGGAATGY-
ACTTGGGAATACC |
|


|
|





















