TCFL2(HMG)/K562-TCF7L2-ChIP-Seq(GSE29196)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACWTCAAAGG
ACWTCAAAGG |
|


|
|
Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 2 |
| Score: | 0.98
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACWTCAAAGG
ACATCAAAGG |
|


|
|
Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer
| Match Rank: | 3 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACWTCAAAGG--
ACATCAAAGGNA |
|


|
|
MA0523.1_TCF7L2/Jaspar
| Match Rank: | 4 |
| Score: | 0.95
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ACWTCAAAGG--
AAAGATCAAAGGAA |
|


|
|
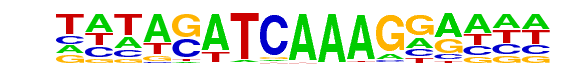
PB0082.1_Tcf3_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.91
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACWTCAAAGG----
TATAGATCAAAGGAAAA |
|

|
|
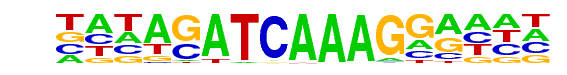
PB0084.1_Tcf7l2_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.91
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ACWTCAAAGG----
NNNAGATCAAAGGANNN |
|

|
|
PB0083.1_Tcf7_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.90
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACWTCAAAGG----
TATAGATCAAAGGAAAA |
|


|
|
PB0040.1_Lef1_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.90
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ACWTCAAAGG----
NANAGATCAAAGGGNNN |
|


|
|
pan/dmmpmm(Bergman)/fly
| Match Rank: | 9 |
| Score: | 0.85
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACWTCAAAGG
AAGATCAAAGG |
|


|
|
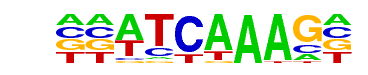
pan/dmmpmm(Papatsenko)/fly
| Match Rank: | 10 |
| Score: | 0.83
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | ACWTCAAAGG
--ATCAAAG- |
|

|
|





















