Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTGTCA
AGGTGTCA |
|


|
|
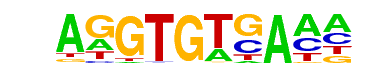
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 2 |
| Score: | 0.92
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTGTCA--
AGGTGTGAAM |
|

|
|
Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer
| Match Rank: | 3 |
| Score: | 0.87
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGGTGTCA--
AGGTGTTAAT |
|


|
|
byn/dmmpmm(Bergman)/fly
| Match Rank: | 4 |
| Score: | 0.85
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTGTCA----
AAGTGCNANAAC |
|


|
|
MA0009.1_T/Jaspar
| Match Rank: | 5 |
| Score: | 0.83
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGGTGTCA-
CTAGGTGTGAA |
|


|
|
PH0164.1_Six4/Jaspar
| Match Rank: | 6 |
| Score: | 0.83
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGGTGTCA-----
TNNNNGGTGTCATNTNT |
|


|
|
PB0013.1_Eomes_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.82
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGGTGTCA-----
GAAAAGGTGTGAAAATT |
|


|
|
E2A-nearPU.1(HLH)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 8 |
| Score: | 0.82
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AGGTGTCA
NNCAGGTGNN- |
|


|
|
PB0117.1_Eomes_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.82
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----AGGTGTCA----
GCGGAGGTGTCGCCTC |
|


|
|
MA0086.1_sna/Jaspar
| Match Rank: | 10 |
| Score: | 0.81
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGGTGTCA
CAGGTG--- |
|


|
|





















