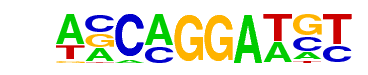
SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACATCCTGNT
ACATCCTGNT |
|


|
|
ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer
| Match Rank: | 2 |
| Score: | 0.86
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACATCCTGNT
ACTTCCGGTT |
|


|
|
EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 3 |
| Score: | 0.85
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACATCCTGNT
ACTTCCTGBT |
|


|
|
ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer
| Match Rank: | 4 |
| Score: | 0.85
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACATCCTGNT
CACTTCCTGT- |
|


|
|
PB0077.1_Spdef_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.85
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ACATCCTGNT----
GTACATCCGGATTTTT |
|


|
|
PB0011.1_Ehf_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.84
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ACATCCTGNT---
TNACTTCCGGNTNNN |
|


|
|
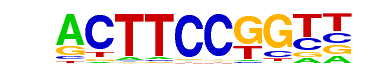
GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer
| Match Rank: | 7 |
| Score: | 0.84
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACATCCTGNT
ACTTCCGGTN |
|

|
|
MA0473.1_ELF1/Jaspar
| Match Rank: | 8 |
| Score: | 0.84
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACATCCTGNT--
CACTTCCTGNTTC |
|


|
|
Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer
| Match Rank: | 9 |
| Score: | 0.84
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ACATCCTGNT
NRYTTCCGGH- |
|


|
|
Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer
| Match Rank: | 10 |
| Score: | 0.84
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ACATCCTGNT
ACTTCCTGTT |
|


|
|