Eomes(T-box)/H9-Eomes-ChIP-Seq(GSE26097)/Homer
| Match Rank: | 1 |
| Score: | 0.63
| | Offset: | 11
| | Orientation: | reverse strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
-----------AGGTGTTAAT |
|


|
|
Tbet(T-box)/CD8-Tbet-ChIP-Seq(GSE33802)/Homer
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 11
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
-----------AGGTGTGAAM |
|


|
|
CG11617/dmmpmm(Noyes_hd)/fly
| Match Rank: | 3 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
ATTTAACATA----------- |
|


|
|
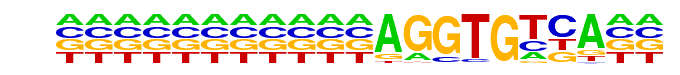
Tbx5(T-box)/HL1-Tbx5.biotin-ChIP-Seq(GSE21529)/Homer
| Match Rank: | 4 |
| Score: | 0.57
| | Offset: | 11
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
-----------AGGTGTCA-- |
|

|
|
PB0013.1_Eomes_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ANTTMRCASBNNNGTGYKAAN
NNTTTTCACACCTTNNN------ |
|


|
|
byn/dmmpmm(Bigfoot)/fly
| Match Rank: | 6 |
| Score: | 0.56
| | Offset: | 11
| | Orientation: | reverse strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
-----------ANGTGCGA-- |
|


|
|
byn/dmmpmm(SeSiMCMC)/fly
| Match Rank: | 7 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
AATTCGCAC------------ |
|


|
|
MA0009.1_T/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | 9
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
---------CTAGGTGTGAA- |
|


|
|
byn/dmmpmm(Bergman)/fly
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ANTTMRCASBNNNGTGYKAAN
AAANTNACACCT---------- |
|


|
|
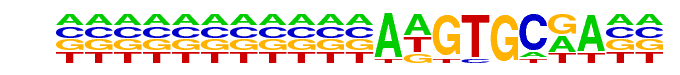
byn/dmmpmm(Pollard)/fly
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 11
| | Orientation: | forward strand |
| Alignment: | ANTTMRCASBNNNGTGYKAAN
-----------AAGTGCGA-- |
|

|
|





















