Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
CCATTGTTNY |
|


|
|
Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer
| Match Rank: | 2 |
| Score: | 0.94
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
CCWTTGTY-- |
|


|
|
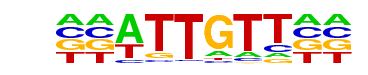
MF0011.1_HMG_class/Jaspar
| Match Rank: | 3 |
| Score: | 0.94
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
--ATTGTT-- |
|

|
|
MA0515.1_Sox6/Jaspar
| Match Rank: | 4 |
| Score: | 0.93
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
CCATTGTTTT |
|


|
|
Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 5 |
| Score: | 0.93
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCATTGTTNY
NCCATTGTTC- |
|


|
|
PB0183.1_Sry_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.92
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CCATTGTTNY----
CNNNTATTGTTCNNNNN |
|


|
|
PB0070.1_Sox30_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.92
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CCATTGTTNY--
ANNTCCATTGTTCNNN |
|


|
|
MA0077.1_SOX9/Jaspar
| Match Rank: | 8 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
CCATTGTTC- |
|


|
|
MA0143.3_Sox2/Jaspar
| Match Rank: | 9 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CCATTGTTNY
CCTTTGTT-- |
|


|
|
MA0445.1_D/Jaspar
| Match Rank: | 10 |
| Score: | 0.91
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CCATTGTTNY
TCCATTGTTCT |
|


|
|





















