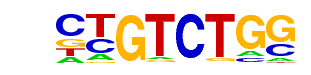
Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTGTCTGG
CTGTCTGG |
|
|
|
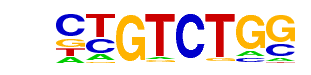
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 2 |
| Score: | 0.98
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --CTGTCTGG
VBSYGTCTGG |
|

|
|
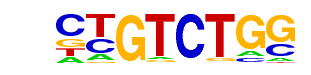
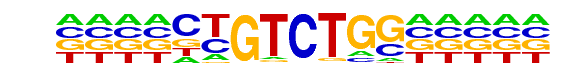
PB0060.1_Smad3_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.94
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CTGTCTGG-----
NNTNNTGTCTGGNNTNG |
|

|
|
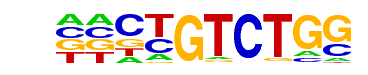
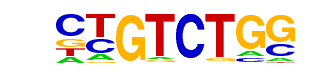
Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer
| Match Rank: | 4 |
| Score: | 0.89
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTGTCTGG
TWGTCTGV |
|

|
|
MA0092.1_Hand1::Tcfe2a/Jaspar
| Match Rank: | 5 |
| Score: | 0.82
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CTGTCTGG---
-GGTCTGGCAT |
|


|
|
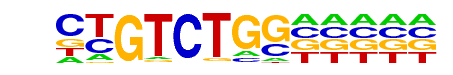
Tbox:Smad/ESCd5-Smad2_3-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 6 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CTGTCTGG-----
-TGTCTGDCACCT |
|

|
|
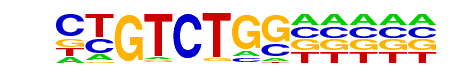
MA0513.1_SMAD2::SMAD3::SMAD4/Jaspar
| Match Rank: | 7 |
| Score: | 0.73
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTGTCTGG-----
CTGTCTGTCACCT |
|

|
|
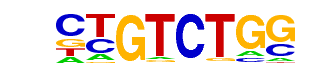
MA0271.1_ARG80/Jaspar
| Match Rank: | 8 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTGTCTGG
GCGTCT-- |
|

|
|
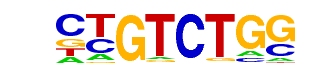
ARG80(MacIsaac)/Yeast
| Match Rank: | 9 |
| Score: | 0.72
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTGTCTGG
GCGTCT-- |
|

|
|
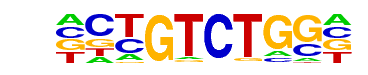
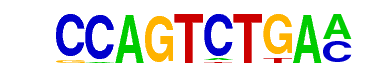
YML081W(MacIsaac)/Yeast
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTGTCTGG-
CCAGTCTGAA |
|
|
|