AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | SCCTSAGGSCAW
SCCTSAGGSCAW |
|


|
|
AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 2 |
| Score: | 0.99
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | SCCTSAGGSCAW
GCCTCAGGGCAT |
|


|
|
MA0003.2_TFAP2A/Jaspar
| Match Rank: | 3 |
| Score: | 0.95
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----SCCTSAGGSCAW
CATTGCCTCAGGGCA- |
|


|
|
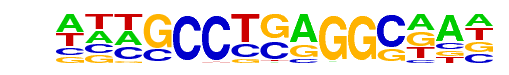
PB0088.1_Tcfap2e_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.94
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---SCCTSAGGSCAW
ATTGCCTGAGGCAAT |
|

|
|
PB0087.1_Tcfap2c_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.93
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---SCCTSAGGSCAW
ATTGCCTGAGGCGAA |
|


|
|
MA0524.1_TFAP2C/Jaspar
| Match Rank: | 6 |
| Score: | 0.92
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----SCCTSAGGSCAW
CATGGCCCCAGGGCA- |
|


|
|
PB0085.1_Tcfap2a_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.91
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---SCCTSAGGSCAW
ATTCCCTGAGGGGAA |
|


|
|
PB0086.1_Tcfap2b_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.84
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --SCCTSAGGSCAW
TTGCCCTAGGGCAT |
|


|
|
PB0190.1_Tcfap2b_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.80
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---SCCTSAGGSCAW
ANTGCCTGAGGCAAN |
|


|
|
PB0189.1_Tcfap2a_2/Jaspar
| Match Rank: | 10 |
| Score: | 0.78
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --SCCTSAGGSCAW
TCACCTCTGGGCAG |
|


|
|





















