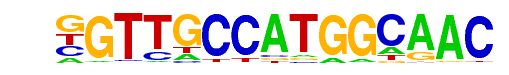
Rfx2(HTH)/LoVo-RFX2-ChIP-Seq(GSE49402)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTTGCCATGGCAACM
GTTGCCATGGCAACM |
|


|
|
RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer
| Match Rank: | 2 |
| Score: | 0.99
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTGCCATGGCAACM-
GTTGCCATGGCAACCG |
|


|
|
RFX1(MacIsaac)/Yeast
| Match Rank: | 3 |
| Score: | 0.97
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTTGCCATGGCAACM
GTTGCCATGGCAAC- |
|


|
|
MA0600.1_RFX2/Jaspar
| Match Rank: | 4 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GTTGCCATGGCAACM----
GTTGCCATGGCAACCGCGG |
|


|
|
MA0509.1_Rfx1/Jaspar
| Match Rank: | 5 |
| Score: | 0.95
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTTGCCATGGCAACM
GTTGCCATGGNAAC- |
|


|
|
Rfx1(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 6 |
| Score: | 0.95
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTTGCCATGGCAACM
-TTGCCATGGCAACN |
|


|
|
X-box(HTH)/NPC-H3K4me1-ChIP-Seq(GSE16256)/Homer
| Match Rank: | 7 |
| Score: | 0.94
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTTGCCATGGCAACM
-TTGCCATGGCAACC |
|


|
|
RFX1/RFX1_YPD/[](Harbison)/Yeast
| Match Rank: | 8 |
| Score: | 0.93
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GTTGCCATGGCAACM
-TTGCCATGGCAAC- |
|


|
|
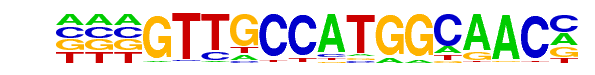
MA0510.1_RFX5/Jaspar
| Match Rank: | 9 |
| Score: | 0.73
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTTGCCATGGCAACM
NCTGTTGCCAGGGAG--- |
|

|
|
PB0055.1_Rfx4_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.72
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GTTGCCATGGCAACM--
--TACCATAGCAACGGT |
|


|
|