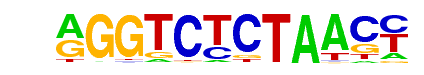
PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCTCTAACC
AGGTCTCTAACC |
|
|
|
NRG1/NRG1_H2O2Hi/[](Harbison)/Yeast
| Match Rank: | 2 |
| Score: | 0.63
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGGTCTCTAACC
-GGACCCT---- |
|

|
|
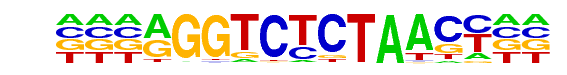
PB0160.1_Rfxdc2_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---AGGTCTCTAACC--
NTNNCGTATCCAAGTNN |
|

|
|
PH0073.1_Hoxc9/Jaspar
| Match Rank: | 4 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGGTCTCTAACC--
GGAGGTCATTAATTAT |
|


|
|
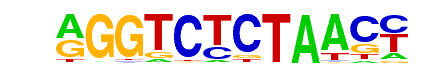
RTG3/Literature(Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGGTCTCTAACC
-GGTCAC----- |
|

|
|
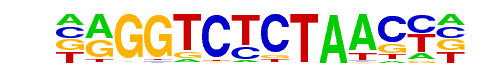
FXR(NR/IR1)/Liver-FXR-ChIP-Seq(Chong et al.)/Homer
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGGTCTCTAACC-
NAGGTCANTGACCT |
|

|
|
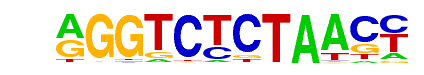
NRG1(MacIsaac)/Yeast
| Match Rank: | 7 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGGTCTCTAACC
-GGACCCT---- |
|

|
|
EcR/usp/dmmpmm(Bergman)/fly
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGGTCTCTAACC--
GAGGTCATTGACCTC |
|


|
|
SPT2/SPT2_YPD/[](Harbison)/Yeast
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -AGGTCTCTAACC
CCTGTCTCTAA-- |
|

|
|
MF0004.1_Nuclear_Receptor_class/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | AGGTCTCTAACC
AGGTCA------ |
|

|
|