PBX1(Homeobox)/MCF7-PBX1-ChIP-Seq(GSE28007)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GSCTGTCACTCA
GSCTGTCACTCA |
|


|
|
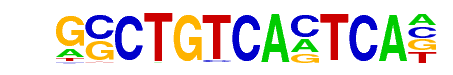
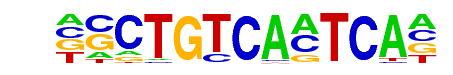
Pbx3(Homeobox)/GM12878-PBX3-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 2 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GSCTGTCACTCA-
-NCTGTCAATCAN |
|
|
|
MA0498.1_Meis1/Jaspar
| Match Rank: | 3 |
| Score: | 0.88
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GSCTGTCACTCA---
AGCTGTCACTCACCT |
|


|
|
PH0141.1_Pknox2/Jaspar
| Match Rank: | 4 |
| Score: | 0.78
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GSCTGTCACTCA
AAGCACCTGTCAATAT |
|


|
|
MA0207.1_achi/Jaspar
| Match Rank: | 5 |
| Score: | 0.77
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GSCTGTCACTCA
--CTGTCA---- |
|


|
|
MA0227.1_hth/Jaspar
| Match Rank: | 6 |
| Score: | 0.75
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GSCTGTCACTCA
--CTGTCA---- |
|


|
|
MA0252.1_vis/Jaspar
| Match Rank: | 7 |
| Score: | 0.75
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GSCTGTCACTCA
--CTGTCA---- |
|


|
|
PH0105.1_Meis3/Jaspar
| Match Rank: | 8 |
| Score: | 0.74
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GSCTGTCACTCA
AATTACCTGTCAATAC |
|


|
|
PH0169.1_Tgif1/Jaspar
| Match Rank: | 9 |
| Score: | 0.74
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GSCTGTCACTCA-
NNNCAGCTGTCAATATN |
|


|
|
PH0140.1_Pknox1/Jaspar
| Match Rank: | 10 |
| Score: | 0.74
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GSCTGTCACTCA
AAAGACCTGTCAATCC |
|


|
|





















