Pax7-long(Paired/Homeobox)/Myoblast-Pax7-ChIP-Seq(GSE25064)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TAATCHGATTAC
TAATCHGATTAC |
|


|
|
Phox2a(Homeobox)/Neuron-Phox2a-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 2 |
| Score: | 0.94
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TAATCHGATTAC
YTAATYNRATTA- |
|


|
|
CES-1(Homeobox)/cElegans-L1-CES1-ChIP-Seq(modEncode)/Homer
| Match Rank: | 3 |
| Score: | 0.78
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TAATCHGATTAC
NAAATTSAATTT- |
|


|
|
PH0126.1_Obox6/Jaspar
| Match Rank: | 4 |
| Score: | 0.75
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TAATCHGATTAC
CNATAATCCGNTTNT |
|


|
|
PH0025.1_Dmbx1/Jaspar
| Match Rank: | 5 |
| Score: | 0.71
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----TAATCHGATTAC
NNNATTAATCCGNTTNA |
|


|
|
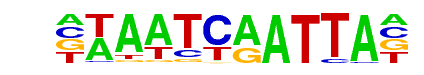
prd/dmmpmm(Papatsenko)/fly
| Match Rank: | 6 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | TAATCHGATTAC
-TAATCAATTA- |
|

|
|
MA0468.1_DUX4/Jaspar
| Match Rank: | 7 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TAATCHGATTAC
TAATTTAATCA- |
|


|
|
Gsc/dmmpmm(Noyes)/fly
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TAATCHGATTAC
TAATCC------ |
|


|
|
MA0190.1_Gsc/Jaspar
| Match Rank: | 9 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | TAATCHGATTAC
TAATCC------ |
|


|
|
MA0467.1_Crx/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -TAATCHGATTAC
CTAATCCTCTT-- |
|


|
|





















