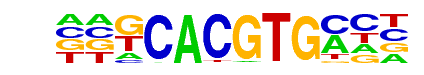bHLHE40(HLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | KCACGTGMCN
KCACGTGMCN |
|


|
|
BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 2 |
| Score: | 0.98
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | KCACGTGMCN
-CACGTGNC- |
|


|
|
MA0281.1_CBF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.97
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | KCACGTGMCN
GCACGTGA-- |
|


|
|
CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 4 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | KCACGTGMCN
-CACGTGDC- |
|


|
|
CBF1/CBF1_SM/72-CBF1(Harbison)/Yeast
| Match Rank: | 5 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | KCACGTGMCN
-CACGTGA-- |
|


|
|
MA0004.1_Arnt/Jaspar
| Match Rank: | 6 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | KCACGTGMCN
-CACGTG--- |
|


|
|
MA0566.1_MYC2/Jaspar
| Match Rank: | 7 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | KCACGTGMCN
GCACGTGC-- |
|


|
|
MA0569.1_MYC4/Jaspar
| Match Rank: | 8 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | KCACGTGMCN
GCACGTGN-- |
|


|
|
MA0568.1_MYC3/Jaspar
| Match Rank: | 9 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | KCACGTGMCN
GCACGTGC-- |
|


|
|
PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer
| Match Rank: | 10 |
| Score: | 0.96
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --KCACGTGMCN
NHBCACGTGV-- |
|
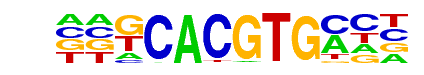

|
|