PAX5(Paired/Homeobox)/GM12878-PAX5-ChIP-Seq(GSE32465)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GCAGCCAAGCGTGACN
GCAGCCAAGCGTGACN |
|


|
|
Pax8(Paired/Homeobox)/Thyroid-Pax8-ChIP-Seq(GSE26938)/Homer
| Match Rank: | 2 |
| Score: | 0.95
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GCAGCCAAGCGTGACN
SCAGYCADGCATGAC- |
|


|
|
MA0014.2_PAX5/Jaspar
| Match Rank: | 3 |
| Score: | 0.93
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----GCAGCCAAGCGTGACN
GAGGGCAGCCAAGCGTGAC- |
|


|
|
MA0069.1_Pax6/Jaspar
| Match Rank: | 4 |
| Score: | 0.74
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GCAGCCAAGCGTGACN
-AANTCATGCGTGAA- |
|


|
|
prd-PD/dmmpmm(Bergman)/fly
| Match Rank: | 5 |
| Score: | 0.66
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | GCAGCCAAGCGTGACN---
--TGTCAACCGTGACGANA |
|


|
|
MA0067.1_Pax2/Jaspar
| Match Rank: | 6 |
| Score: | 0.64
| | Offset: | 8
| | Orientation: | reverse strand |
| Alignment: | GCAGCCAAGCGTGACN
--------NCGTGACN |
|


|
|
prd/dmmpmm(Noyes)/fly
| Match Rank: | 7 |
| Score: | 0.63
| | Offset: | 7
| | Orientation: | reverse strand |
| Alignment: | GCAGCCAAGCGTGACN----
-------AGCGTGACGGATT |
|


|
|
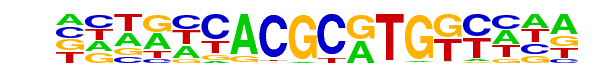
PL0008.1_hlh-29/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | GCAGCCAAGCGTGACN--
-CTGCCACGCGTGGCCAA |
|

|
|
MA0006.1_Arnt::Ahr/Jaspar
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | GCAGCCAAGCGTGACN
-------TGCGTG--- |
|


|
|
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | 9
| | Orientation: | forward strand |
| Alignment: | GCAGCCAAGCGTGACN
---------CATGAC- |
|


|
|





















