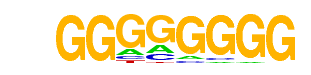
Maz(Zf)/HepG2-Maz-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | GGGGGGGG
GGGGGGGG |
|
|
|
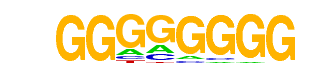
MA0599.1_KLF5/Jaspar
| Match Rank: | 2 |
| Score: | 0.88
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -GGGGGGGG-
GGGGNGGGGC |
|


|
|
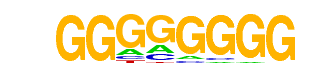
SeqBias: polyC-repeat
| Match Rank: | 3 |
| Score: | 0.87
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GGGGGGGG--
GGGGGGGGGG |
|

|
|
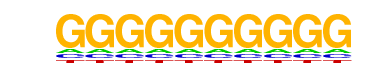
PB0100.1_Zfp740_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.86
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----GGGGGGGG---
NANNTGGGGGGGGNGN |
|


|
|
MA0039.2_Klf4/Jaspar
| Match Rank: | 5 |
| Score: | 0.83
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -GGGGGGGG-
TGGGTGGGGC |
|


|
|
MA0079.3_SP1/Jaspar
| Match Rank: | 6 |
| Score: | 0.83
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --GGGGGGGG-
GGGGGCGGGGC |
|


|
|
PB0097.1_Zfp281_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.83
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GGGGGGGG----
GGGGGGGGGGGGGGA |
|


|
|
PB0107.1_Ascl2_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.83
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GGGGGGGG----
NATNGGGNGGGGANAN |
|


|
|
PB0039.1_Klf7_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.82
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GGGGGGGG----
NNAGGGGCGGGGTNNA |
|


|
|
POL003.1_GC-box/Jaspar
| Match Rank: | 10 |
| Score: | 0.79
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---GGGGGGGG---
AGGGGGCGGGGCTG |
|


|
|