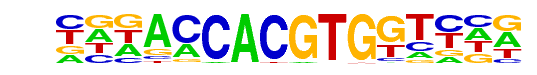Max(HLH)/K562-Max-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
ACCACGTGGTNN |
|


|
|
c-Myc/mES-cMyc-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 2 |
| Score: | 0.98
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ACCACGTGGTNN
-CCACGTGGNN- |
|


|
|
n-Myc(HLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 3 |
| Score: | 0.98
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ACCACGTGGTNN
-CCACGTGGNN- |
|


|
|
MA0568.1_MYC3/Jaspar
| Match Rank: | 4 |
| Score: | 0.97
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
-GCACGTGC--- |
|


|
|
BMAL1(HLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 5 |
| Score: | 0.97
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
GNCACGTG---- |
|


|
|
c-Myc(HLH)/LNCAP-cMyc-ChIP-Seq(unpublished)/Homer
| Match Rank: | 6 |
| Score: | 0.97
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
NCCACGTG---- |
|


|
|
PL0007.1_mxl-3/Jaspar
| Match Rank: | 7 |
| Score: | 0.96
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ACCACGTGGTNN-
CGGACCACGTGGTCCG |
|

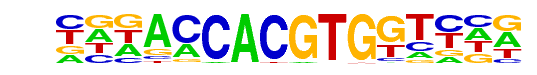
|
|
CLOCK(HLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 8 |
| Score: | 0.96
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
GHCACGTG---- |
|


|
|
PHO4/PHO4_Pi-/38-PHO4(Harbison)/Yeast
| Match Rank: | 9 |
| Score: | 0.96
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | ACCACGTGGTNN
--CACGTGG--- |
|


|
|
PIF5ox(bHLH)/Arabidopsis-PIF5ox-ChIP-Seq(GSE35062)/Homer
| Match Rank: | 10 |
| Score: | 0.96
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ACCACGTGGTNN
NHBCACGTGV--- |
|


|
|