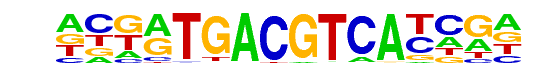c-Jun-CRE(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATGACGTCATCN
ATGACGTCATCN |
|


|
|
JunD(bZIP)/K562-JunD-ChIP-Seq/Homer
| Match Rank: | 2 |
| Score: | 0.99
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATGACGTCATCN
ATGACGTCATCN |
|


|
|
MA0492.1_JUND_(var.2)/Jaspar
| Match Rank: | 3 |
| Score: | 0.94
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATGACGTCATCN--
NATGACATCATCNNN |
|


|
|
PB0038.1_Jundm2_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.94
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATGACGTCATCN-
CCGATGACGTCATCGT |
|


|
|
Atf1(bZIP)/K562-ATF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 5 |
| Score: | 0.93
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ATGACGTCATCN
-TGACGTCATC- |
|


|
|
PB0004.1_Atf1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.92
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATGACGTCATCN-
ACGATGACGTCATCGA |
|

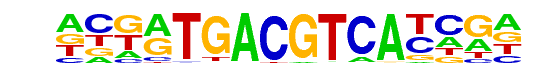
|
|
MA0488.1_JUN/Jaspar
| Match Rank: | 7 |
| Score: | 0.91
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGACGTCATCN-
ATGACATCATCNN |
|


|
|
STF1(bZIP)/Glycine max/AthaMap
| Match Rank: | 8 |
| Score: | 0.88
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATGACGTCATCN
NATGACGTCATC- |
|


|
|
MA0018.2_CREB1/Jaspar
| Match Rank: | 9 |
| Score: | 0.87
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ATGACGTCATCN
-TGACGTCA--- |
|


|
|
CRE(bZIP)/Promoter/Homer
| Match Rank: | 10 |
| Score: | 0.85
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATGACGTCATCN
GTGACGTCACCG |
|


|
|