Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | MTGATGCAAT
MTGATGCAAT |
|


|
|
Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer
| Match Rank: | 2 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | MTGATGCAAT
ATGATGCAAT |
|


|
|
AARE(HLH)/mES-cMyc-ChIP-Seq/Homer
| Match Rank: | 3 |
| Score: | 0.95
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | MTGATGCAAT-
-TGATGCAATC |
|


|
|
CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 4 |
| Score: | 0.89
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -MTGATGCAAT
NATGTTGCAA- |
|


|
|
MA0466.1_CEBPB/Jaspar
| Match Rank: | 5 |
| Score: | 0.84
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | MTGATGCAAT-
ATTGTGCAATA |
|


|
|
MA0488.1_JUN/Jaspar
| Match Rank: | 6 |
| Score: | 0.81
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---MTGATGCAAT
AAGATGATGTCAT |
|


|
|
MF0006.1_bZIP_cEBP-like_subclass/Jaspar
| Match Rank: | 7 |
| Score: | 0.81
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | MTGATGCAAT
-TTATGCAAT |
|


|
|
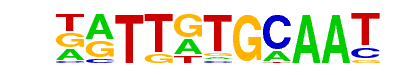
MA0102.3_CEBPA/Jaspar
| Match Rank: | 8 |
| Score: | 0.79
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -MTGATGCAAT
NATTGTGCAAT |
|

|
|
MA0492.1_JUND_(var.2)/Jaspar
| Match Rank: | 9 |
| Score: | 0.74
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----MTGATGCAAT-
AAAGATGATGTCATC |
|


|
|
MA0019.1_Ddit3::Cebpa/Jaspar
| Match Rank: | 10 |
| Score: | 0.73
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | MTGATGCAAT---
-AGATGCAATCCC |
|


|
|





















