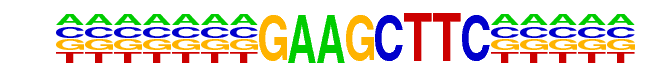HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 1 |
| Score: | 1.00
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
NNTTCTGGAANNTTCTAGAA |
|


|
|
HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer
| Match Rank: | 2 |
| Score: | 0.91
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
-----TAGAANVTTCTAGAA |
|


|
|
MA0486.1_HSF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.89
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
-CTTCTAGAAGGTTCT---- |
|


|
|
HSF1(MacIsaac)/Yeast
| Match Rank: | 4 |
| Score: | 0.85
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
------GGAACGTTCTAGAA |
|


|
|
Hsf/dmmpmm(Pollard)/fly
| Match Rank: | 5 |
| Score: | 0.78
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
-----TCGAAACTTCTAGA- |
|


|
|
SFL1(MacIsaac)/Yeast
| Match Rank: | 6 |
| Score: | 0.75
| | Offset: | 7
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
-------GAAGCTTC----- |
|

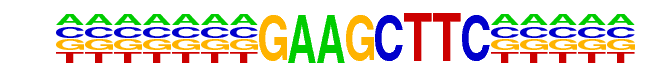
|
|
HSF1/HSF1_H2O2Lo/11-HSF1(Harbison)/Yeast
| Match Rank: | 7 |
| Score: | 0.71
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
--TTCTAGNNCNTTC----- |
|


|
|
PB0090.1_Zbtb12_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.60
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA---
------CTAAGGTTCTAGATCAC |
|


|
|
shn/dmmpmm(Papatsenko)/fly
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
-----GGGAACGTTCCC--- |
|


|
|
TaMYB80(MYB)/Triticum aestivum/AthaMap
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | 6
| | Orientation: | reverse strand |
| Alignment: | NNTTCTGGAANNTTCTAGAA
------NGGATATTCCC--- |
|


|
|