
HOMER
Software for motif discovery and ChIP-Seq analysis
Functional Enrichment Analysis
HOMER contains a program for performing functional
enrichment analysis
from a list of Entrez Gene IDs (findGO.pl).
Normally,
you
don't
need
to know how to use findGO.pl
because it is called
internally by findMotifs.pl
and annotatePeaks.pl.
findGO.pl assesses
the enrichment
of various categories of gene function, biological pathways,
domain
structure, chromosome location, etc., in your gene list
relative to a
set of background gene IDs. Enrichment is calculated
assuming the
cumulative hypergeometric
distribution, much in the same way that HOMER scores
motif
enrichment. HOMER does not attempt to deal with the
multiple-hypothesis testing problem, although it does record
the number
tests made in each output file.There are several different "ontologies", or libraries of gene groupings, that HOMER will check for enrichment. Below is the list, sources in "()"s:
- Biological Process: Functional groupings of proteins (Gene Ontology)
- Molecular Function: Mechanistic actions of proteins (Gene Ontology)
- Cellular Component: Protein localization (Gene Ontology)
- Chromosome Location: Genes with similar chromosome
localization (NCBI Entrez Gene)
- KEGG Pathways: Groups of proteins in the same pathways (From KEGG)
- Protein-Protein Interactions: Groups of proteins interacting with the same protein (From NCBI Entrez Gene)
- Interpro: Proteins with similar domains and features (Interpro)
- Pfam: Proteins with similar domains and features (Pfam)
- SMART: Proteins with similar domains and features (SMART)
- Gene3D: Proteins with similar domains and features (Gene3D Database)
- Prosite: Proteins with similar domains and features (Prosite Database)
- PRINTS: Proteins with similar domains and features (PRINTS Database)
- MSigDB: Lists of genes maintained by the Molecular Signature Database (includes many different categories of genes (MSigDB)
- BIOCYC: Groups of proteins in the same pathway (NCBI Biosystems/BIOCYC)
- COSMIC: Human proteins that are mutated in the same cancers (COSMIC)
- GWAS Catalog: Human genes with risk SNPs identified in their vicinity for the same disease (GWAS Catalog)
- Lipid Maps: Mouse proteins found in the same lipid processing pathways (NCBI Biosystems/LIPID MAPS)
- Pathway Interaction Database: Proteins in the same pathway (NCBI Biosystems/PID)
- REACTOME: Proteins in the same biochemical pathways (NCBI Biosystems/REACTOME)
- SMPDB: Proteins in the same pathway (SMPDB)
- Wikipathways: Protein in the same pathway
(Wikipathways)
To run findGO.pl on its own, type:
findGO.pl
<input
file
of
Entrez
Gene IDs> <organism> <output
directory> [-bg <background ID file>] [-cpu #]
[-human]
There are a couple of newer options for findGO.pl that can also be triggered through findMotifs.pl:
-bg <background Gene File> : By default HOMER will use the *.base.gene file found in the homer/data/promoters/ directory for background, which normally represents all gene IDs for the organism. You can use this option to specify a specific background.
-cpu <#> : number of CPUs/threads to use for GO analysis (each ontology will be given it's own thread)
-human : Use Homologene to first convert IDs to human (can useful for non-model organisms) - only way to check COSMIC/GWAS groups if in another organism.
Normally findGO.pl will use a default set of gene ids for that organism. The program produces one HTML file, containing a mixture of different enriched categories, as well tab-delimited text files for each of the ontologies analyzed. An example of the GO output is show below:
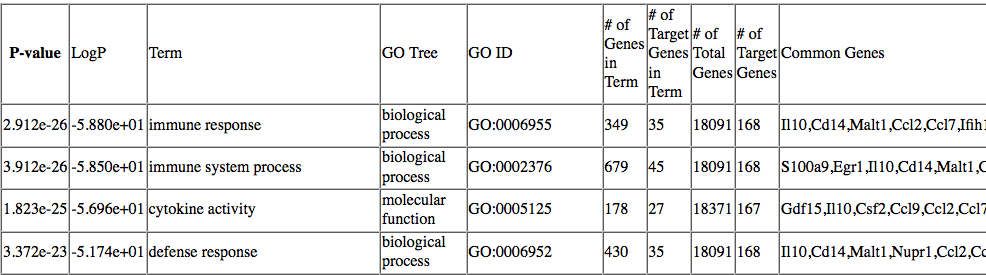
In the HTML page, findGO.pl
will
convert the gene IDs into gene symbols so that it is easy
to
read. In the text files the IDs are kept as gene
IDs.
Gene Ontology and GO slims
My favorite topic in the
world of
Gene Ontology analysis is the use of GO slims.
HOMER does not
contain GO slims libraries. As a result, you may
find that many
of your gene ontology results contain terms such as "metabolism" and "cellular process"
when other tools
may not reveal these terms. GO slims are great
because they
delete terms that you don't generally want to see.
Another way to
do this is to look through your list and just use the
terms you
want. There really isn't much of a difference
between that and
using GO slims - but at least
you're being honest with yourself with one of the
techniques.

Can't figure something out? Questions, comments, concerns, or other feedback:
cbenner@ucsd.edu